May 5 2017
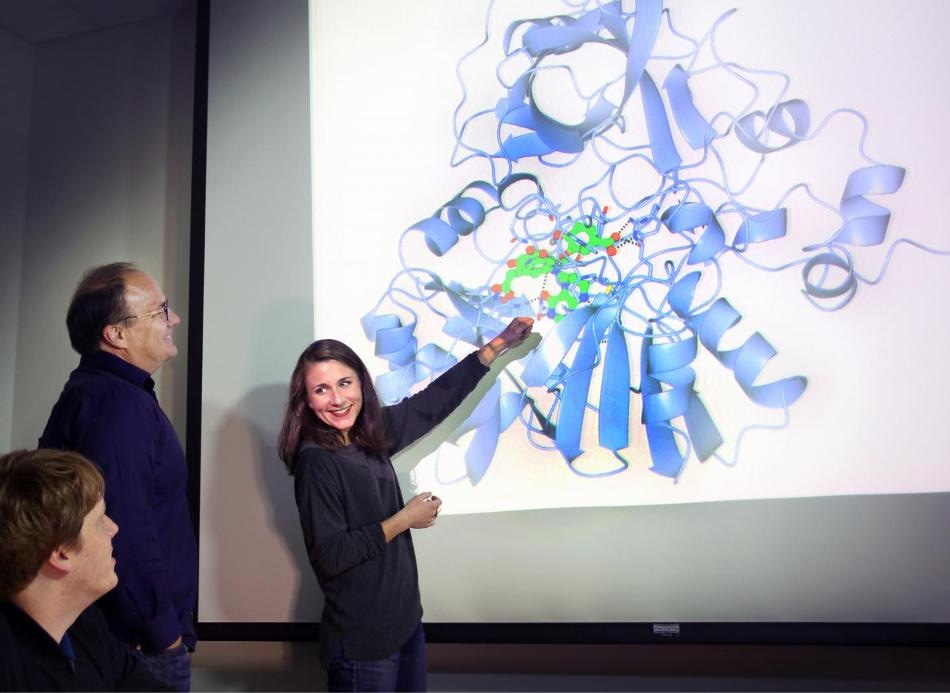 Sandia National Laboratories researchers Matthew Mills, left, and Ken Sale, center, look on as researcher Amanda Kohler points toward a model of LigM, an enzyme whose structure they have solved. (Photo by Dino Vournas)
Sandia National Laboratories researchers Matthew Mills, left, and Ken Sale, center, look on as researcher Amanda Kohler points toward a model of LigM, an enzyme whose structure they have solved. (Photo by Dino Vournas)
Sandia National Laboratories researchers may have unlocked the potential of biofuel waste, which would eventually make biofuels competitive with petroleum, with their recent discovery.
Fuel produced from plants is a lot more expensive than petroleum; however one way to lower the cost would be to sell products made from lignin, the left over plant waste from biofuel production.
Lignin usually is either burned to create electricity or left unused in piles because there are no solutions yet to determine how to convert it into useful products, such as nylon, fabrics, renewable plastics, and adhesives. The electricity is not even available to the general public; it is only used by companies that generate large quantities of lignin, like pulp and paper manufacturers. Now Sandia researchers, collaborating with researchers from Lawrence Berkeley National Laboratory at the Joint BioEnergy Institute, have decoded the structure and behavior of LigM, an enzyme that breaks down molecules derived from lignin.
The team’s study on LigM appears in the most recent issue of the Proceedings of the National Academy of Sciences. The enzyme has little in common with other, better understood proteins, which formerly made it difficult for researchers to deduce how it works. This paper marks the first time a research has solved the structure of LigM, paving the way toward new molecules and new, saleable products.
Revealing nature’s closely-guarded secrets
For years, researchers have struggled with the issue of breaking down lignin, the part of plant cell walls that gives structure and protection from insect and bacterial attacks. This strength also makes lignin hard to deconstruct, though there have been fresh breakthroughs.
The plant matter used to create ethanol can be physically or chemically pre-treated so that the lignin is deconstructed in the process. However, these techniques can be expensive and decrease the quantity of biofuel that can be harvested. They could also obstruct later-stage lignin harvesting. That is why a few researchers are concentrating on discovering enzymes that convert lignin gently and naturally.
Sandia researcher and lead author Amanda Kohler said her team understood that enzymes could metabolize lignin and its derivatives because there are age-old records of bacteria using enzymes for this reason. Sphingomonas bacteria were discovered living in the waste water of a pulp mill over three decades ago. Once researchers comprehended that the bacterium’s unique enzymatic pathways enabled it to live on lignin, their task then was to examine the enzymes in these pathways so they could imitate what nature had already accomplished, and use that understanding effectively.
Enzymatic pathway to profit
Kohler and her team concentrated on LigM, an enzyme used by Sphingomonas, as it performs a main step in the conversion of lignin derivatives and it is the simplest of the known enzyme systems that does this function.
When trying to mimic natural systems in a laboratory setting, the simplest, most direct systems are the best.
Amanda Kohler, Researcher, Sandia National Laboratories
The team discovered that half of LigM’s structure is made up of a common protein architecture present in all forms of life, from humans to bacteria. The rest of the enzyme — the active portion — is not present in any other identified protein structure. This unique structure offers LigM the capacity to bind specifically to molecules derived from lignin.
Solving the structure allows us to understand how the organism may have evolved its unique function, which I think is scientifically one of the most interesting findings.
Ken Sale, Scientist, Sandia national Research
The team used the Advanced Light Source Synchrotron at Lawrence Berkeley National Laboratories, along with high-performance computing and fundamental biochemistry to achieve their insights into LigM.
LigM is built to only break down lignin derivatives, not lignin itself. It is essential to understand that LigM’s work is only one main step in a longer pathway of reactions required to totally deconstruct lignin. One active area of research includes discovering other organisms, possibly fungi that can perform the first step of breaking down large lignin mass into smaller bits. Some of the Sandia researchers, Sale and Matthew Mills, who solved LigM’s structure, have now learned more about another enzyme that helps trigger the breakdown of lignin into smaller bits.
LigM acts on a later stage in the process, when smaller lignin bits already have been converted into a molecule called vanillic acid.
There is still work to be done to figure out the whole reaction pathway. But now we have a much-needed understanding of a key step in this process and are developing enzymes to fit our end goals of lowering the cost of biofuels by making products from lignin.
Amanda Kohler, Researcher, Sandia National Laboratories