Nov 14 2017
New research offers an insight into a unique enzyme that could provide an eco-friendly treatment for chlorite-contaminated water supplies and enhance water quality around the world.
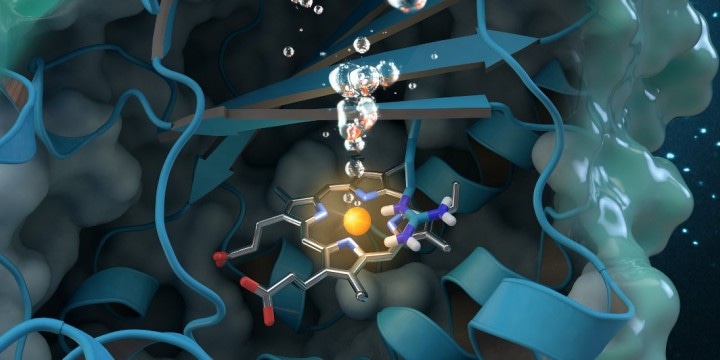 CREDIT - Journal cover art reprinted with permission from ACS Catalysis, vol. 7, issue 11, November 3, 2017. Further permissions related to the material excerpted should be directed to the American Chemical Society.
CREDIT - Journal cover art reprinted with permission from ACS Catalysis, vol. 7, issue 11, November 3, 2017. Further permissions related to the material excerpted should be directed to the American Chemical Society.
A global team of researchers headed by Christian Obinger from the University of Vienna used neutron analysis at Oak Ridge National Laboratory, X-ray crystallography and other methods to analyze the chlorite dismutase enzyme. This naturally occurring protein can break down chlorite, into harmless byproducts, but its catalytic process is not properly understood. Chlorite is an industrial pollutant found in groundwater, soils, and drinking water. Comprehending how the bacterial enzyme converts chlorite into oxygen and chloride could pave the way for future applications in biotechnology and bioremediation.
The results, published in ACS Catalysis, also contribute to core research on the enzyme’s ability to create oxygen. Oxygen generation is extremely rare in nature, once thought feasible only by photosynthesis, so the enzymatic activity of chlorite dismutase has gained a lot of interest from the scientific community beyond its environmental applications for clean water.
Precisely how chlorite dismutase functions at a molecular level to break down chlorite has been discussed since the enzyme was found in 1996. The intricacy of the enzyme’s molecular structure and the difficulty of examining proteins with experimental approaches pose inherent challenges for scientists.
Similar to a majority of enzymes, chlorite dismutase is a protein that catalyzes a very specific reaction. The process is mostly environmentally dependent, meaning it works best within specific parameters, including concentration, temperature, and pH ranges. Identifying the suitable parameters for the reaction is paramount to supporting bioengineering and mass production of chlorite dismutase to safely eliminate chlorite from the environment and potentially make the most of the enzyme’s oxygen generation.
The team isolated an unexplored Cyanothece strain of chlorite dismutase and analyzed the protein’s crystal structure at specific pH values to establish the influence of pH on chlorite conversion.
The researchers used MaNDi, the macromolecular neutron diffractometer, beamline 11-B at the Spallation Neutron Source, a Department of Energy User Facility at ORNL, to gather unique data only attainable through the use of neutrons.
Different protein crystals have different degrees of symmetry, which will determine how we go about measuring them. This crystal is unusual in that it has very little symmetry, so an especially large number of reflections have to be recorded individually to get a complete data set, this would be a challenging and lengthy task anywhere, and it was only achievable in this time frame due to the large area detector coverage of the MaNDi instrument.
Leighton Coates, MaNDi Lead Instrument Scientist.
On MaNDi, the researchers were able to spot the protonation states of vital amino acids believed to support the reaction. “Protonation” refers to a central step in catalysis during which hydrogen attaches to molecules. “This is the important region of the protein, where the chemistry is happening and the chlorite is being broken down,” said Coates.
Protonation states are not easily detected because they involve hydrogen, which is hard to spot with X-rays or other methods. Furthermore, a phenomenon called “photoreduction” occurs when exposing metal-containing enzymes like chlorite dismutase to X-rays, fundamentally altering the atomic structure of the sample.
As neutron methods do not have these restrictions, they can provide researchers vital information that cannot be gained using other methods.
Neutrons are nondestructive and sensitive to light elements like hydrogen, so they can provide exclusive information about the atomic structure of proteins, which are largely composed of hydrogen molecules, and unlike x-rays that can damage delicate proteins, neutron techniques allow you to collect data at room temperature on an unaltered protein in its active state without the impacts of ionizing radiation and photoreduction, this experiment really highlights the benefit of using neutrons to study proteins.
Leighton Coates, MaNDi Lead Instrument Scientist.
The journal article is published under the title “Molecular mechanism of enzymatic chlorite detoxification: insights from structural and kinetic studies.” Coauthors include Irene Schaffner, Georg Mlynek, Nicola Flego, Dominic Pühringer, Julian Libiseller-Egger, Leighton Coates, Stefan Hofbauer, Marzia Bellei, Paul G. Furtmüller, Gianantonio Battistuzzi, Giulietta Smulevich, Kristina Djinović-Carugo and Christian Obinger.
The Spallation Neutron Source is a DOE Office of Science User Facility. UT-Battelle manages ORNL for the DOE Office of Science. The single largest supporter of basic research in the physical sciences in the United States, the Office of Science aims to address some of the most pressing challenges currently facing mankind.