Mar 18 2021
Researchers have performed genome surgery of a single-celled alga to eliminate unnecessary parts. This research can prove to be a most efficient cellular factory for generating sustainable biofuels from carbon dioxide and sunlight.
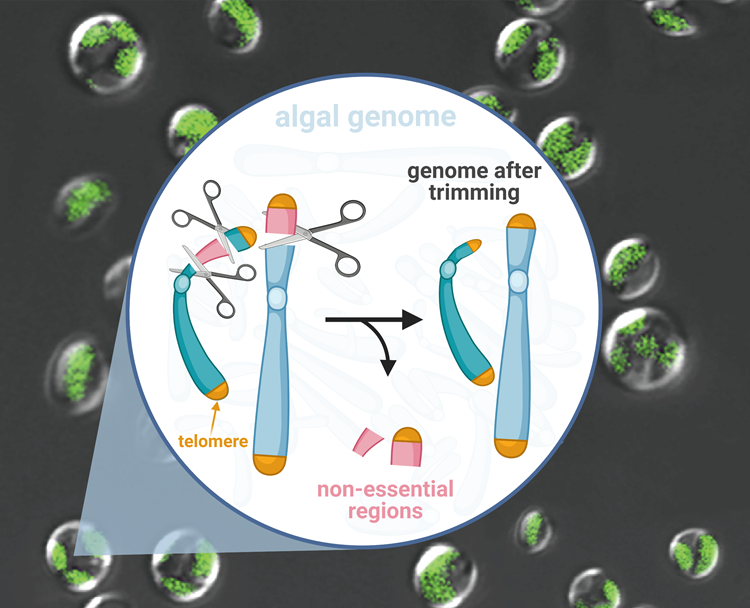 Hundred-kilobase fragment deletions in microalgae by Cas9 cleavages. This figure was made using BioRender. Image Credit: Yang Liu.
Hundred-kilobase fragment deletions in microalgae by Cas9 cleavages. This figure was made using BioRender. Image Credit: Yang Liu.
At the Qingdao Institute of BioEnergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences (CAS), scientists have stripped 100-kilobase genome from a kind of oil-producing microalgae, eliminating genes that are not needed for it to work. Thus, they have developed a 'genome scalpel' that can trim microalgal genomes creatively and quickly.
The 'minimal genome' microalgae made by the team is prospectively beneficial as a model organism for further analysis of the biological and molecular function of each gene, or as a 'chassis' strain for synthetic biologists to supplement for tailored production of biomolecules like bioplastics or biofuels.
The research was reported on March 14th, 2021, in The Plant Journal.
It can be highly beneficial to develop a 'minimal genome'—a genome from which all apparently non-functional or duplicated 'junk genes' —for examining basic questions regarding genetic function and for developing cell factories that generate useful compounds.
These minimal genomes have been made for simple organisms, but hardly for eukaryotic organisms, such as plants or algae. In the case of higher eukaryotes, 'junk' regions can constitute up to 70% of the genome.
Removing what only seems to be 'junk genes' can, in fact, have detrimental impacts on the organism or even kill it.
Scientists from QIBEBT have generated, for the first time, a genome with targeted deletions of 100 kilobases each in size, for a type of algae known as Nannochloropsis oceanica.
N. oceanica are microalgae (single-celled algae) with an enormous potential for producing biomaterials, biofuels and other platform chemicals in a sustainable and renewable manner while decreasing greenhouse gas emissions.
But comprehensive genetic engineering of the organism is required to tap the potential of such microalgae to optimize yields and reduce production costs.
Initially, the QIBEBT team determined the chromosomal regions that are unwanted—the ones whose genes were hardly expressed or activated. Ten such 'low expression regions' or LERs were determined by the researchers.
Then, CRISPR-Cas9 gene-editing technique was used to cut out two of the biggest LERs—which weigh more than 200 kilobases in size.
Despite the all snipping, the microalgae still showed essentially normal growth, lipid content, fatty acid saturation levels and photosynthesis. In some cases, there was even a slightly higher growth rate and biomass productivity than the organism in the wild.
Qintao Wang, Study First Author, Single-Cell Center, Qingdao Institute of BioEnergy and Bioprocess Technology, Chinese Academy of Sciences
“We interestingly found normal telomeres in the telomere-deletion mutants of Chromosome 30. This phenomenon implies the losing of distal part of chromosome may induce telomere regeneration,” stated Jian Xu, the corresponding author of the study from the SCC in QIBEBT.
The considerably snipped genome should already act as a closer-to-minimal genome in Nannochloropsis, which has the ability to act as the chassis strain for tailored production of biomolecules with the help of additional metabolic engineering on top of this chassis.
As the researchers have now proven the genome of such a complicated eukaryote can be stripped down, they want to analyze whether additional LERs and other non-lethal regions can be snipped, to craft a completely minimal Nannochloropsis that produces biofuels from CO2 with the greatest efficiency.
Journal Reference:
Wang, Q., et al. (2021) Genome engineering of Nannochloropsis with hundred‐kilobase fragment deletions by Cas9 cleavages. The Plant Journal. doi.org/10.1111/tpj.15227.